[Recent works]
I. Studies on the dynamics of chromosomes/chromatin fibers, the mechanism of higher-order structural transitions, and their applications
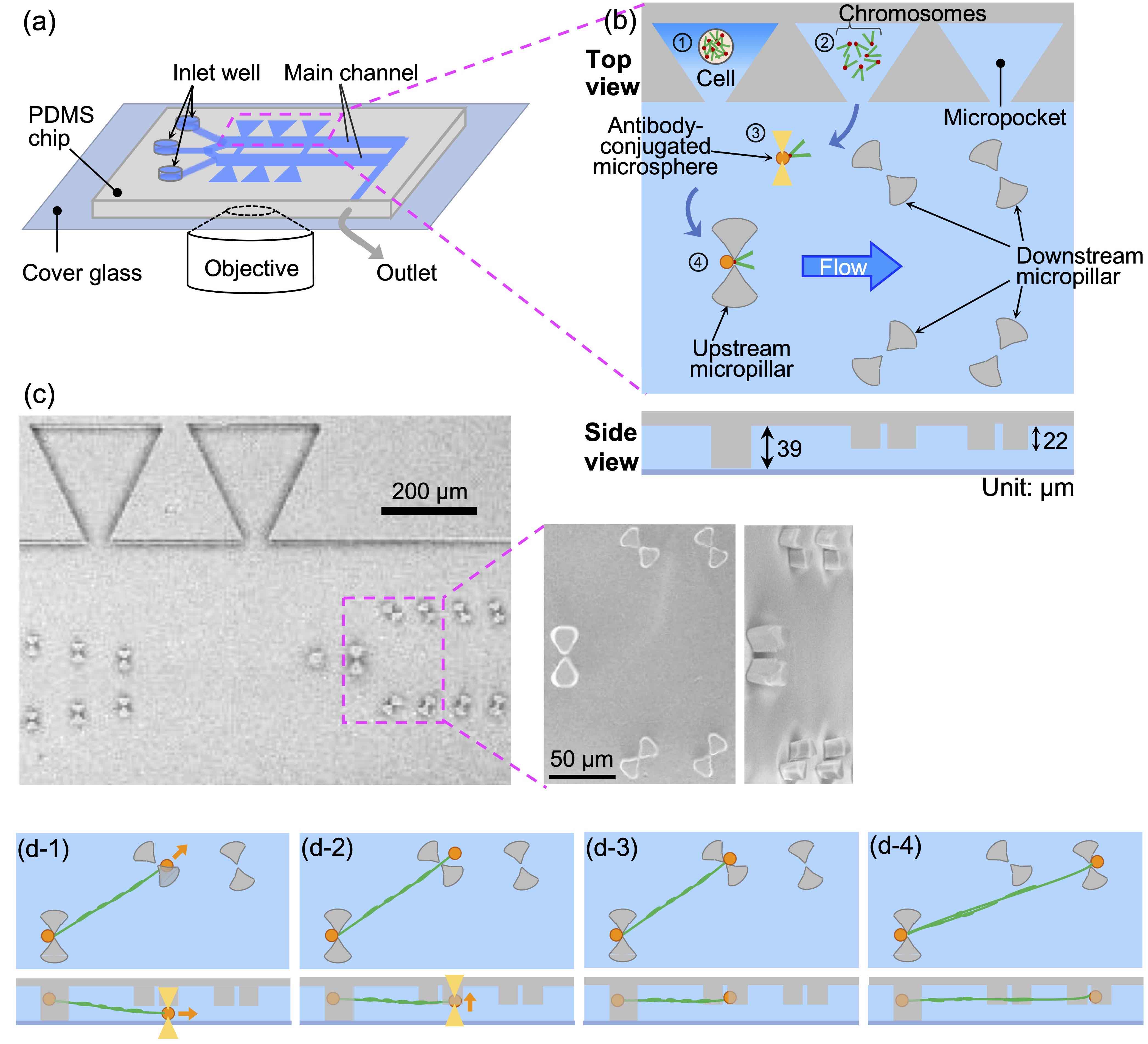
Toward single-cell epigenome analysis: A microfluidic device for isolating, stretching, and imaging individual chromosomesSensors and Actuators B: Chemical, 394, Article number: 134462 (7 pages) (2023)
Abstract: We developed a microfluidic device with microstructures that can moor the ends of chromatin fibers obtained by unfolding single intact chromosomes. This mooring involves trapping both ends of the chromosome with antibody-conjugated microspheres and placing each microsphere on the microstructure. In principle, the device is structured in such a way that mooring of chromatin fibers, i.e., partially unfolded chromosomes, is achieved under uniform tension, which is necessary to obtain an accurate distribution of folded/unfolded regions and fluorescence probes on the chromatin fibers. In addition, changes in the distribution of chromatin folding structures were observed by controlling the solution conditions, and specific histone chemical modification regions were visualized by immunofluorescence staining of chromatin fibers in the elongated form with uniform tension. This method is advantageous for the epigenetic analysis of non-fragmented natural chromatin fibers.
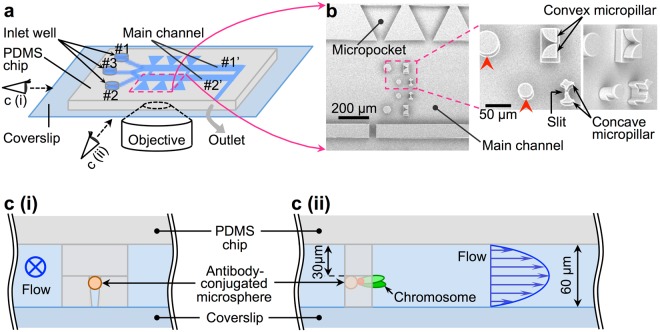
A microfluidic device for isolating intact chromosomes from single mammalian cells and probing their folding stability by controlling solution conditions
Scientific Reports, 8, Article number: 13684 (10 pages) (2018)
Abstract:
Chromatin folding shows spatio-temporal fluctuations in living undifferentiated cells, but fixed spatial heterogeneity in differentiated cells. However, little is known about variation in folding stability along the chromatin fibres during differentiation. In addition, effective methods to investigate folding stability at the single cell level are lacking. In the present study, we developed a microfluidic device that enables non-destructive isolation of chromosomes from single mammalian cells as well as real-time microscopic monitoring of the partial unfolding and stretching of individual chromosomes with increasing salt concentrations under a gentle flow. Using this device, we compared the folding stability of chromosomes between non-differentiated and differentiated cells and found that the salt concentration which induces the chromosome unfolding was lower (≤500 mM NaCl) for chromosomes derived from undifferentiated cells, suggesting that the chromatin folding stability of these cells is lower than that of differentiated cells. In addition, individual unfolded chromosomes, i.e., chromatin fibres, were stretched to 150–800 µm non-destructively under 750 mM NaCl and showed distributions of highly/less folded regions along the fibres. Thus, our technique can provide insights into the aspects of chromatin folding that influence the epigenetic control of cell differentiation.
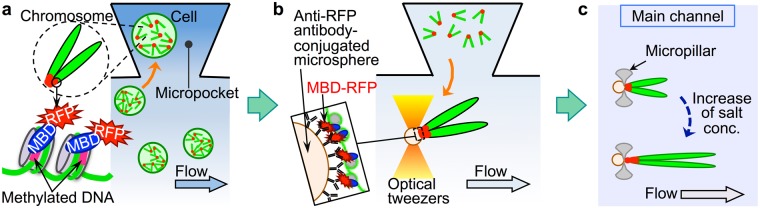
Nucleosomes Exhibit Non-uniform Unwrapping Along Native Chromatin Fibers with Increasing Salt Concentration as Revealed by Direct Imaging in a Microfluidic Channel
Biotechnology Journal, 13, 1700245 (8 pages) (2018)
Abstract:
Identifying the distribution of the higher-order structure of chromatin - a complex of DNA and proteins - along genomic DNA can clarify the mechanisms underlying cell development and differentiation, including gene regulation. However, genome-wide analysis of this distribution at the single-cell level remains an outstanding challenge. Here, the authors report a new method for investigating changes in and the distribution of higher-order structures along native chromatin fibers - ranging over 100 µm in length - relative to changes in salt concentration. To this end, the authors developed a microfluidic platform that enabled us to isolate chromatin fibers from single cells and tether them to microstructures in a microfluidic channel without fragmentation. The fibers were then exposed to varying concentrations of salt solution under microscopic observation. As a result, the fibers are non-uniformly elongated by up to 2-3 times along the fiber axis as salt concentration was increased from 0 to 3 M, suggesting that chromosome structural stability is non-uniformly distributed along chromatin fibers in their native form. Thus, our system enables direct microscopic analysis of individual chromatin fibers from single cells, which can provide insights into epigenetic mechanisms of cell development, cell differentiation, and carcinogenesis. (C) 2017 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
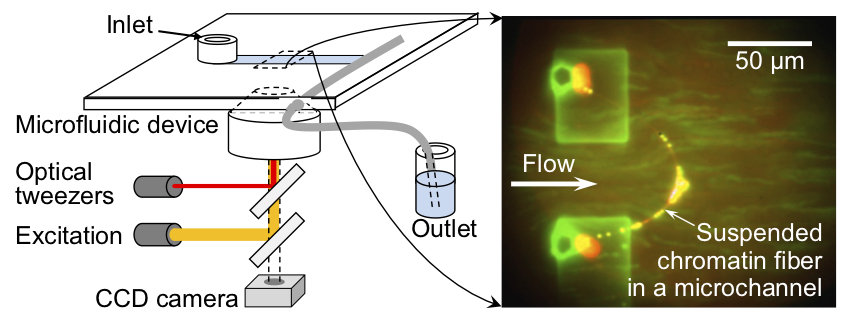
Non-destructive handling of individual chromatin fibers isolated from single cells in a microfluidic device utilizing an optically driven microtool
Lab on a Chip, 14, 696-704 (2014)
Abstract:
We report a novel method for the non-destructive handling of, and biochemical experiments with, individual intact chromatin fibers, as well as their isolation from single cells, utilizing a specifically designed microfluidic device with an optically driven microtool under the microscope. With the aid of the microtool, isolated chromatin fibers were handled non-destructively, and were tethered at the microstructures fabricated in the microfluidic device with straightened conformation by the flow. Immunofluorescence staining was carried out as a demonstrative biochemical experiment with the individual native chromatin fibers isolated in the microfluidic device. The potential application of this method for epigenetic analyses of intact chromatin fibers isolated from single cells is demonstrated. (C) Royal Society of Chemistry 2014.
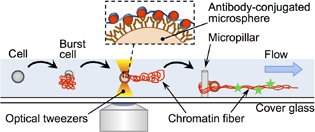
II. Studies on interactions between DNA and proteins and their application
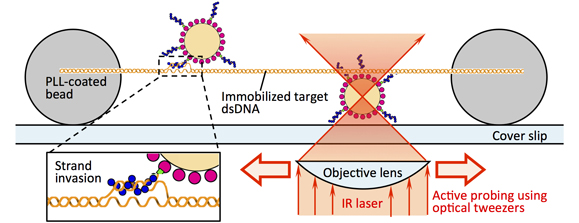
Real-Time FISH using Optically-driven Microspheres Functionalized by the Homologous Recombination Protein, RecAProc. 16th International Conference on Miniaturized Systems for Chemistry and Life Sciences (MicroTAS 2012), 770-772 (2012)
Abstract: Here, we demonstrate the active probing of a specific sequence without thermal denaturation of a target double-stranded DNA by using a recombination protein, RecA. A microsphere with single-stranded DNA-RecA complexes on its surface was optically manipulated and slid onto the target DNA, and a strong interaction between the target DNA and the microspheres at the specific position of the target DNA was observed. This result shows the potential use of recombination proteins to facilitate detection of specific DNA sequences with spatial information, i.e., real-time FISH. (C) 2012 Chemical and Biological Microsystems Society.
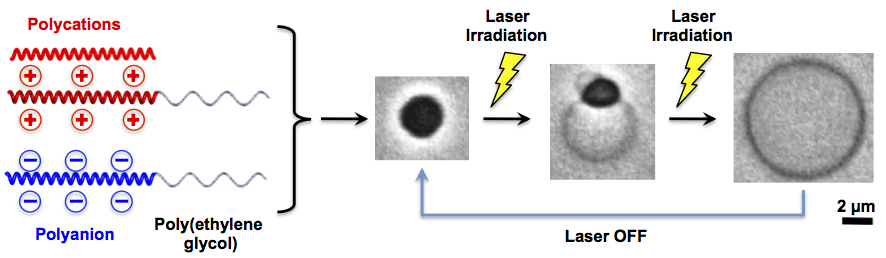
III. Studies on polymer vesicles
Direct Formation of Giant Unilamellar Vesicles from Microparticles of Polyion Complexes and Investigation of Their Properties Using a Microfluidic ChamberSoft Matter, 9, 5448-5458 (2013)
Abstract: Herein, a novel approach is demonstrated for organic-solvent-free preparation of giant unilamellar vesicles utilizing the unique response of polyion complexes (PICs) to changes in additive salt concentration. A microfluidic device consisting of a main channel bearing side pockets that work as microscale reaction chambers is designed for facilitating the preparation process under an optical microscope. With this device, real-time observation of morphological transformation of individual PIC microparticles is carried out during rapid reduction of the additive salt concentration and direct formation of giant vesicles from PIC microparticles is shown. (C) Royal Society of Chemistry 2013.

Spontaneous Formation of Giant Unilamellar Vesicles from Microdroplets of Polyion Complex via Thermally Induced Phase Separation
Angewandte Chemie International Edition, 48, 4613-4616 (2009)
Abstract: Herein, we report the spontaneous formation of giant
unilamellar polyion complex (PIC) vesicles from micronsized
PIC coacervates that are based on biocompatible
polyelectrolyte precursors. Vesicle formation is demonstrated
in pure aqueous solution under thermal perturbation, which
was achieved by irradiation with a focused infrared laser from
an optical tweezer setup. We also describe how this morphological
transformation is developed through thermally
induced generation and growth of a water-rich phase inside
the PIC coacervate. Additionally, the PIC coacervate acts as a
“thermally driven pump” that transports water from the
solution outside the vesicle to the water-rich phase within
(i.e., to balloon the PIC coacervate for the formation of a PIC
vesicle). (C) 2009 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
IV. Development of cell function modification technology/device
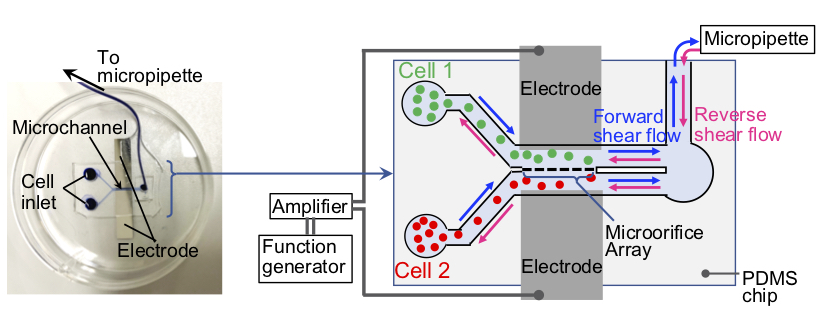
A single-cell surgery microfluidic device for transplanting tumor cytoplasm into dendritic cells without nuclei mixingBiotechnology Journal, e2200135 (9 pages) (2023)
Abstract: This study aimed to demonstrate the feasibility of generating tumor cell vaccine models by single-cell surgery in a microfluidic device that integrates one-to-one electrofusion, shear flow reseparation, and on-device culture. The device was microfabricated from polydimethylsiloxane (PDMS) and consisted of microorifices (aperture size: ∼3 μm) for one-to-one fusion, and microcages for on-device culture. Using the device, we could achieve one-to-one electrofusion of leukemic plasmacytoid dendritic cells (DClike cells) and Jurkat cells with a fusion efficiency of ∼ 80%. Fusion via the narrow microorifices allowed DC-like cells to acquire cytoplasmic contents of the Jurkat cells while preventing nuclei mixing. After fusion, the DC-like cells were selectively reseparated from the Jurkat cells by shear flow application to generate tumor nuclei-free antigen-recipientDC-like (tarDC-like) cells.When cultured as single cells on the device, these cells could survive under gentle medium perfusion with a median survival time of 11.5 h, although a few cells could survive longer than 36 h. Overall, this study demonstrates single-cell surgery in a microfluidic device for potential generation of dendritic cell vaccines which are uncontaminated with tumor nucleic materials. We believe that this study will inspire the generation of safer tumor cell vaccines for cancer immunotherapy.
[Previous works]
1) Dynamics of large DNA molecules during gel electrophoresis
H. Oana, Y. Masubuchi, M. Matsumoto, M. Doi, Y. Matsuzawa, K. Yoshikawa,
Periodic Motion of Large DNA Molecules during Steady Field Gel Electrophoresis,
Macromolecules, 27, 6061-6067 (1994).
Abstract:
Direct observation of individual T4 DNA molecules during steady field gel electrophoresis was carried out with fluorescence microscopy. Statistical analyses of the image data show that (i) DNA molecules change their conformation and velocity periodically even under a steady field, (ii) the characteristic time of the periodic motion decreases with an increase of the electric field and with a decrease of the gel concentration, and (iii) the rear end of DNA shows a periodic stick-slip motion, while the front end moves with almost constant speed. A possible interpretation for the periodic behavior is presented.
2) Injection process of DNA from solution to gel under steady field
H. Oana, Y. Masubuchi, M. Matsumoto, M. Doi, K. Yoshikawa,
Motion of Large DNA Molecules Traveling from Solution to Gel Under Steady Field,
Journal of Polymer Science: Part B: Polymer Physics, 34, 1105-1111 (1996).
Abstract:
Direct observation of individual T4 DNA molecules at the interface between gel and pure solvent under steady electric field was carried out with fluorescence microscopy. Statistical analyses of the image data show that (1) when a DNA arrives at the gel surface, it remains there for a short time before the DNA stretches its arms to enter the gel, and (2) the time spent on the gel surface increases with an increase of the agarose persentage in the gel. (C) 1996 John Wiley& Sons, Inc.
3) Dynamics of large DNA molecules during electrophoresis in Concentrated Polymer Solutions
H. Oana, M. Doi, M. Ueda, K. Yoshikawa,
Reorientation of Large DNA Molecules in Concentrated Polyacrylamide Solution during Crossed Field Electrophoresis,
Electrophoresis, 18, 1912-1915 (1997).
Abstract:
Recently, we found that, in concentrated neutral solutions, DNA molecules migrate in linear conformation under steady electric field. In this paper, we report the conformational change of DNA during 120 degrees crossed-field electrophoresis in the same polymer solution. We found that, in concentrated polyacrylamide solutions, the reorientation process of DNAs becomes simple: the DNA goes back along the previous track and the reorientation time is longer for larger DNA. Such a backtrack motion has been thought to be an essential motion for the separation of DNA fragments in pulsed field gel electrophoresis. We expect that this phenomenon is useful for a more efficient separation technique of large DNAs than the current pulsed field gel electrophoresis.
4) High speed separation of linear and supercoiled DNA by CE
H. Oana, R. W. Hammond, J. J. Schwinefus, S.-C. Wang, M. Doi, M. D. Morris,
High Speed Separation of Linear and Supercoiled DNA by Capillary Electrophoresis. Buffer, Entangling Polymer and Electric Field Effects,
Analytical Chemistry, 70, 574-579 (1998).
Abstract:
Capillary electrophoresis in dilute and semidilute (slightly entangled) hydroxyethyl cellulose (HEC) is shown to separate linear double-stranded DNA (ds-DNA) and supercoiled plasmid DNA in the size range 1-16 thousand base pairs in 3 min, The mobilities of linear ds-DNA fragments are stronger functions of electric field strength and buffer concentration than the mobilities of supercoiled plasmids, The effects of HEC concentration and molecular weight are similar for both forms of DNA, The behavioral differences, which are attributed to the greater stiffness of the plasmids, can be used to define conditions that maximize resolution of supercoiled and linear ds-DNA of the same or similar number of base pairs.
5) Developing a method for single molecule analysis of large DNA
H. Oana, M. Ueda, and M. Washizu,
Visualization of a Specific Sequence on a Single Large DNA Molecule Using Fluorescence Microscopy Based on a New DNA-Stretching Method,
Biochem. Biophys. Res. Commun., 265, 140-143 (1999).
Abstract:
A method for analyzing large DNA which makes it possible to obtain spatial information on the positions of specific sequences along a DNA molecule has been developed. Making use of the fact that large DNA molecules are stably elongated under an alternating-current field in a concentrated linear polymer solution, the direct observation of elongated individual lambda DNA molecules with fluorescence probes was carried out using fluorescence microscopy. The spatial positions of the fluorescent spots of the probe (fluorescence-labeled restriction endonuclease EcoRI) on DNA molecules were determined by image analysis. As expected, fluorescent spots of EcoRI were observed at certain positions on lambda DNA, where sequences to which EcoRI binds are located. Finally, the potential application of single large DNA molecule analysis using this DNA-stretching method is discussed. (C) 1999 Academic Press.
6) Study on the relation between the biochemical characteristics of DNA and its higher-order structure
H. Oana, K. Tsumoto, Y. Yoshikawa, K. Yoshikawa,
Folding transition of large DNA completely inhibits the action of a restriction endonuclease as revealed by single-chain observation,
FEBS Lett., 530, 143-146 (2002).
Abstract:
The biochemical characteristics of lambda DNA chains in folded/unfolded states upon cleavage by the restriction enzyme ApaLI were investigated in the presence of spermine. These characteristics of DNA chains depending on their higher-order structure were studied at the single-molecule level using fluorescence microscopy. With a low concentration of spermine, lambda DNA takes a random coiled conformation and allows digestion by the enzyme, while under a high concentration of spermine, lambda DNA takes a compact folded structure and inhibits such attack. Together with comparative experiments on short oligomeric DNA, our results suggest that the transition in the higher-order structure causes on/off-type switching of sensitivity to the enzyme. (C) 2002 Published by Elsevier Science B.V. on behalf of the Federation of European Biochemical Societies.
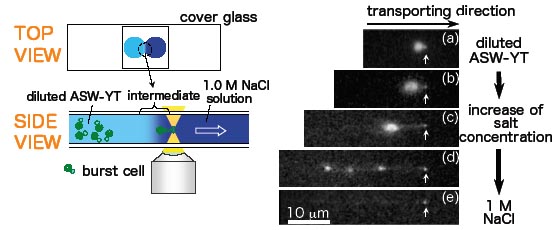
7) On-site manipulation of single whole-genome DNA molecules recovered from single cells
H. Oana, K. Kubo, K. Yoshikawa, H. Atomi, and T. Imanaka,
On-site manipulation of single whole-genome DNA molecules using optical tweezers,
Appl. Phys. Lett., 85, 5090-5092 (2004).
Abstract:
In this report, we describe a noninvasive methodology for manipulating single Mb-size whole-genome DNA molecules. Cells were subjected to osmotic shock and the genome DNA released from the burst cells was transferred to a region of higher salt concentration using optical tweezers. The transferred genome DNA exhibits a conformational transition from a compact state into an elongated state, accompanied by the change in its environment. The applicability of optical tweezers to the on-site manipulation of giant genome DNA is suggested, i.e., lab-on-a-plate. (C) 2004 American Institute of Physics.
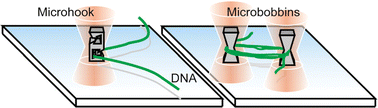
8) Individual manipulation of large DNA molecules by optically-driven microtools
K. Terao, M. Washizu and H. Oana,
On-site manipulation of single chromosomal DNA molecules by using optically driven microstructures
Lab on a Chip, 8, 1280-1284 (2008)
Abstract: We report a novel method for manipulation of single giant DNA molecules under a video microscope. Using optically driven microstructures, we manipulated chromosomal DNA of length in the order of millimeters, extended by electroosmotic flow without DNA breakage in aqueous solution: we picked up DNA, using microfabricated hooks and wound it around microfabricated bobbins. (C) Royal Society of Chemistry 2008.
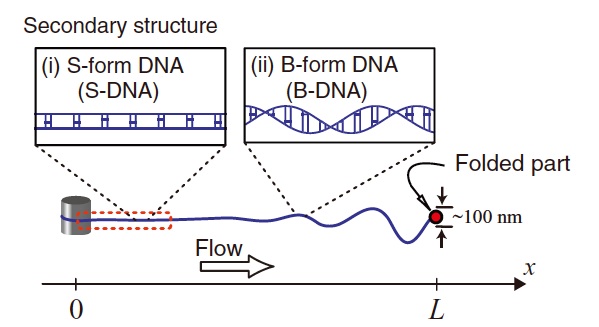
9) Folding dynamics of tethered giant DNA under strong flow in the presence of a condensing agent
T. Saito, T. Sakaue, D. Kaneko, M. Washizu, and H. Oana,
Folding dynamics of tethered giant DNA under strong flow
J. Chem. Phys. 135, 154901 (2011)
Abstract: Using a microfluidic device, we investigate the folding dynamics of individual linear long DNA, whose one end is tethered under a strong flow in the presence of a condensing agent. Direct observations of the folding process of DNA molecules reveal a characteristic dynamics with pronounced non-monotonic velocity of the folded part at the free end against the flow.We discuss this unique dynamics in relation to the inhomogeneous spatial fluctuation and the structure change at the multiple order levels along the stretched DNA, which is induced by the increasing tension due to the build-up of the hydrodynamic drag force. (C) 2011 American Institute of Physics.